
There is a whole menagerie of variations on the Southern's original theme: Far-Western blots, Far-Eastern blots, Middle Eastern blots, Southwestern blots etc. Many different techniques have been variously described as Eastern blotting, most of which involve using different types of probes to detect post-translational modifications of proteins, for instance the use of lectins to probe a protein-blotted membrane in search of specific carbohydrates. Like Southerns, Western blots are also used for diagnostic assays, for instance, the confirmation of HIV infection.Īs the only direction remaining, the term "Eastern blotting" has been fought over extensively. The proteins are then transferred onto a membrane and probed with specific antibodies. Proteins are separated by one or two-dimensional polyacrylamide gel electrophoresis-usually by isoelectric point in one dimension, and by size, in the presence of denaturing detergent, in the other direction. The pun was stretched even further to include Western blots, also developed in the Stark laboratory.
NORTHERN HYBRIDIZATION FREE
Blotbase, from , is a free database of mRNA sequences quantified by Northern blots from various cell types and tissues. This is particularly important in problems of gene regulation, such as embryogenesis and the concomitant cell differentiation. Northern blots were invaluable for detecting and quantifying specific mRNAs, thus determining the activity of different genes. Molecular biologists being punny fellows, this was immediately dubbed the Northern blot. Very rapidly, a similar technique was developed to analyze RNA by James Alwine, David Kemp, and George Stark at Stanford. Southern's technique has literally become an industry. A number of prisoners have been released from Death Row on the basis of DNA assays performed years after their original trial. Now, of course, the Southern blot has also been adapted for forensic DNA analysis. "Isn't that great?" enthused Tony Hunter, the Salk Institute scientist sitting next to me, clearly overjoyed (inasmuch as his English reserve would allow him) that molecular biology lab techniques were being used for something so obviously medically useful. Not long after, while still a graduate student, I attended a seminar wherein the investigator demonstrated the use "Southern blots" to diagnose genetic diseases. Overnight, sales of paper towels to molecular biology labs skyrocketed. The membrane could then be analyzed with labeled DNA probes that would hybridize to specific sequences. The DNA was first treated with restriction enzymes to make the fragments, then subjected to agarose gel electrophoresis to separate them by size, then denatured in situ and transferred to a nitrocellulose membrane by virtue of sucking volumes of buffer through the gel and the membrane, using stacks of paper towels to blot up the buffer. Thus the reverse procedure, though originally uncommon, enabled the one-at-a-time study of gene expression using northern analysis to evolve into gene expression profiling, in which many (possibly all) of the genes in an organism may have their expression monitored.In 1975, British biologist Edwin Southern published a method for detecting specific DNA fragments after separating them by size. The use of DNA microarrays that have come into widespread use in the late 1990s and early 2000s is more akin to the reverse procedure, in that they involve the use of isolated DNA fragments affixed to a substrate, and hybridization with a probe made from cellular RNA. In this procedure, the substrate nucleic acid (that is affixed to the membrane) is a collection of isolated DNA fragments, and the probe is RNA extracted from a tissue and radioactively labelled. For smaller fragments denaturing polyacrylamide urea gels are employed.Īs in the Southern blot, the hybridization probe may be made from DNA or RNA.Ī variant of the procedure known as the reverse northern blot was occasionally (although, infrequently) used. A notable difference in the procedure in case of agarose gels, (as compared with the Southern blot) is the addition of formaldehyde which acts as a denaturant. The gels may be run on either agarose or denaturing polyacrylamide gels depending on the size of the RNA to be detected. This technique was developed in 1977 by James Alwine, David Kemp, and George Stark at Stanford University.
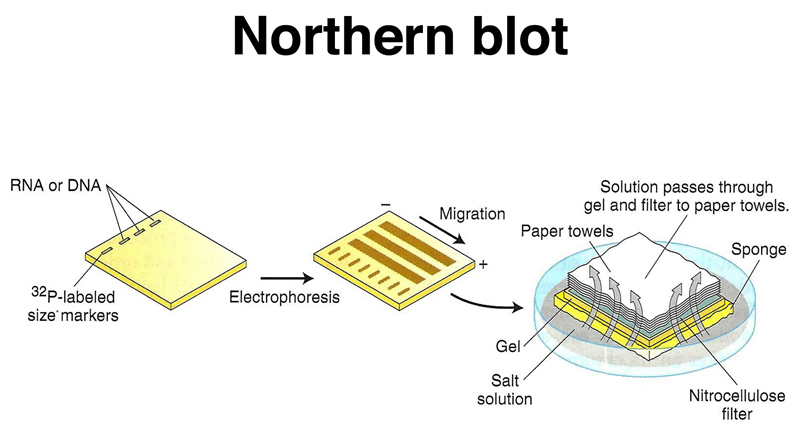
It takes its name from the similarity of the procedure to the Southern blot procedure, named for biologist Edwin Southern, used to study DNA, with the key difference that, in the northern blot, RNA, rather than DNA, is the substance being analyzed by electrophoresis and detection with a hybridization probe. The northern blot is a technique used in molecular biology research to study gene expression.


 0 kommentar(er)
0 kommentar(er)
